Overview
My research centers on the development of new statistical methodology for neural networks, network modeling, clustering and model selection,
with applications to high-dimensional biological data.
My group consists of 3 PhD students and 3 postdocs. We work on projects ranging from multi-group and multi-view
data integration, multilayer network modeling, interpretable deep neural networks and analysis challenges in systems biology. For more details, please check the List of Current Projects below - we are always looking
for master students who want to get involved in research projects.
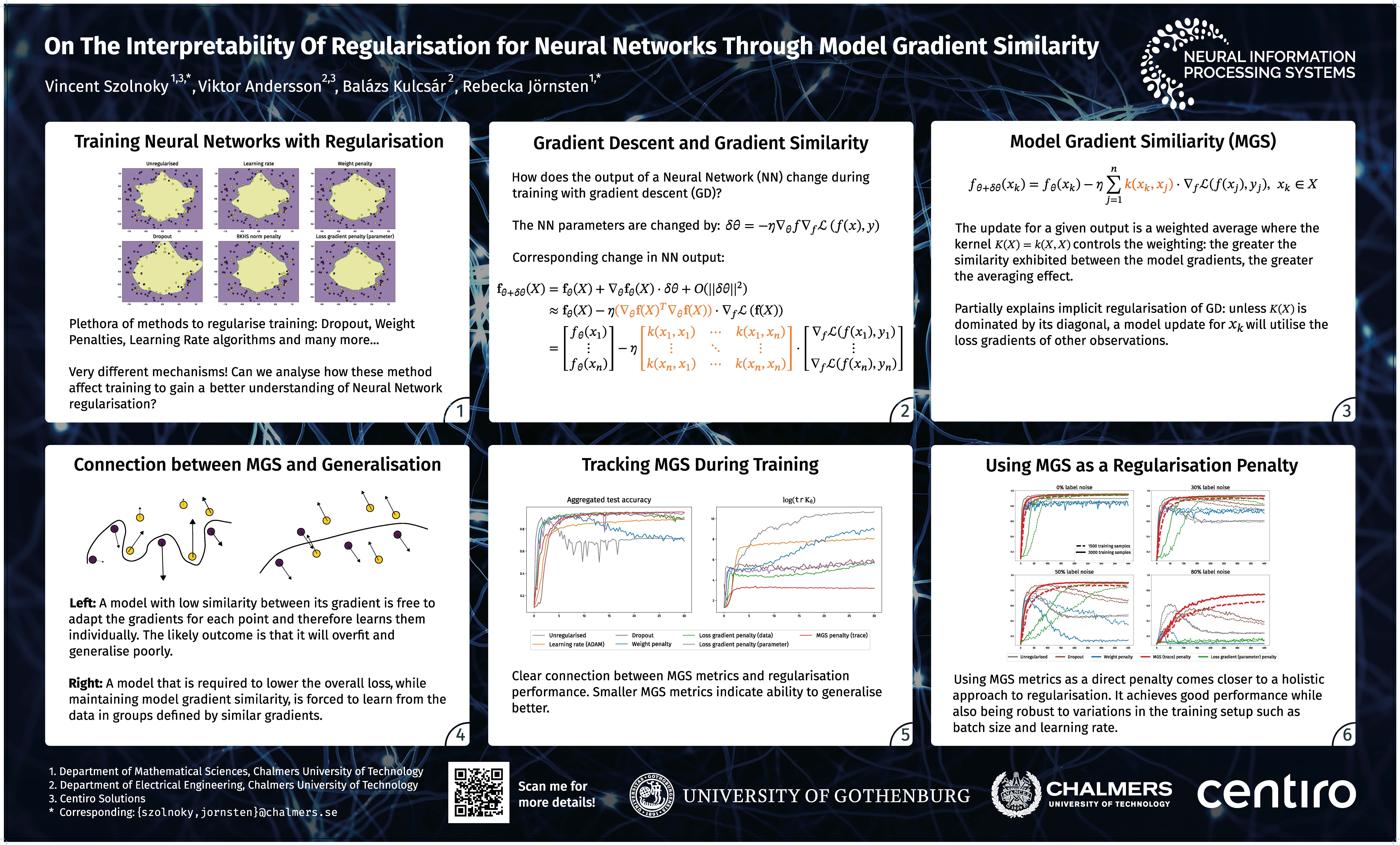
Data integration and joint modeling are a rich source for research problems. These types of problems are central components in
several joint projects in collaboration with Sven Nelander's group.
Our group aims to formulate integrated models for mRNA, microRNA, DNA copy number, methylation and mutation in human cancer. These projects are supported
by two grants from the Swedish Foundation for Strategic Research as well as the Wallenberg AI, Autonomous Systems and Software
Program (WASP)
and the SciLifeLab and Wallenberg National Program for Data-Driven Life Science (DDLS).
We also collaborate with Mika Gustafsson's group at
Linköping university on data integration tasks, with a special emphasis on the utilization of prior biological information. This project is also funded through WASP and DDLS.
I collaborate with Johan Jonasson on new regularization techniques
for deep neural networks to better handle the presence of mislabeled data. This
project is supported by the Wallenberg foundation
In a joint research project with
Professor Giuseppe Durisi , Chalmers Electrical Engineering, we investigate the generalization properties of deep neural networks -
how come such over-parameterized models perform so well on test data?
The project "Generalization bounds for Deep Neural Networks: Insight and Design" is funded through the WASP Graduate School .
Together with Balazs Kulcsar at
Electrical Engineering, we work on research problems merging control theory and neural networks.
Centiro, have funded two industry PhD positions on this topic.
Since 2018, I am the Vice-Dean for Research and Research Infrastructures at Faculty of Science , University of Gothenburg.
I chair the Faculty Board's advisory committee, where we discuss interdepartmental activities, review sabbatical applications and
Faculty prizes and awards. We also organize faculty events.
List of Current Projects
We are always looking for master students who want to be a part of our research team. Please contact me if any of the projects sound interesting to you.
You are also welcome to contact me with your own project idea. I usually supervise 5-7 master students each year.
AI, Deep Learning
Neural Networks and Control Theory: Together with Professor Balazs Kulcsar at Electrical Engineering, I supervise PhD students Viktor Andersson and Vincent Szolnoky, funded by
Centiro . In this team we work on problems ranging from methodological and theoretical challenges in neural networks to practical applications in logistics and transportation.
AI-Integromics: We have several theses projects centering on the development of data integration methods for systems biology using
deep neural networks paired with structured regularization. 2 postdocs in this group work in this area in collaboration with Mika Gustafsson's group at
Linköping university and Sven Nelander's group at Uppsala University.
Machine Learning, Systems Biology
Flexible Joint Matrix Factorization: My PhD student Felix Held is working on
both theoretical and practical aspects of data integration through matrix factorization. There are many open problems one can pursue here; scalability, applications, generalizations.
Accelerated training and regularization of neural networks: My PhD students Oskar Allerbo and Maria Matveev work on
methodological projects centering on new regularization techniques and training strategies for neural networks.
Teaching Philosophy
My classes are usually made up of a mix of students; undergraduates, master students and PhD students and all from different fields of study. I enjoy this kind of dynamic classroom.
I tend to mix black-board lectures with computer demonstrations for all my classes. My goal in teaching is that the students leave my class recognizing that statistical modeling is not a "push-the-button" type exercise, and every data set requires unique consideration.
Courses & Workshops
In past years I have cycled teaching applied statistics couses (Linear models, Applied multivariate analysis) and method courses (Statistical inference principles and Survival analysis).
My main focus for teaching now is Statistical Learning for Big Data and master thesis projects.
Here are some links to recent courses:
People
Alums
- Jonatan Kallus 2014-2020 Integrative modeling of cancer.
- Jose Sanchez, 2009-2014 Network models with applications to genomic data: generalization, validation and uncertainty assessment Currently working as an analyst at Astra Zeneca
- Alexandra Jauhiainen 2009-2010 Statistics in Gene Expression, Metabolomics, and Comparative Genomics in Evolution (co-supervisor). Currently working as principal statistician at Astra Zeneca
Current Master students
- Maria Matveev Accelerated training of neural networks using coordinated learning
- Anton Eliasson Gustafsson and Kayed Mahra. Modeling delays in maritime transport, project with Centiro.
- Gustav Johannesson and Alexander Lindhardt Machine Learning Based Charging Decision
Policy for a Fleet of Electric Vehicles , project with Volvo
Alums
- 2021: Only examiner commitments due to faculty administration
- 2020: Selma Tabakovic, Oskar Liew, Per Ed.
- 2019: Jacob Söderström, Isak Hjortgren, Sandra Larsson, Jacob Lindbäck, Mikael Böörs, Elijah Ferreira, Arash Shahsavari, Sandra Vikander, Linn Engström, Andreas Syren, Viktor Andersson, Natalja Vessilinova, Elijah Ferrera, Wade Rosco
- 2018: Olof Ekborg, Philip Arndt, Fredrik Beiron, Johan Björk, Adam Brinkman, Fredrik Kjernald, Karl Svensson.
- 2017: Andrew Nisbet, Leif Schelin, Alex Bergkvist, Sebastian Franzen, Filip Birve
- 2016: Björn Hedner, Nils Wireklint, Emilio Jorge, Pasha Hashemi, Ludvig Vikström, Oskar Lilja, Maja Fahlen
- 2015: Sofia Hjalmarsson, Sebastian Anerud, Susanne Pettersson, Linus Lundin
- 2002-2014: Johanna Svensson, Patrik Johansson, Viktor Skokic, Johanna Sigmundsdottir, Tobias Abenius, Eric Burlow, Owen Martin, Diane Richardson